Difference between revisions of "Evoptool: Evolutionary Optimization Tool"
(→Evoptool package) |
(→Download) |
||
| Line 145: | Line 145: | ||
==== Wrapped Algorithms ==== | ==== Wrapped Algorithms ==== | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 15:30, 7 March 2010
Contents
Project profile
| Name: | Evoptool: Evolutionary Optimization Tool. |
| Field: | Evolutionary Computation (EC). |
| People involved: | Matteo Matteucci - matteucci@elet.polimi.it
Luigi Malagò - malago@elet.polimi.it Gabriele Valentini - gabriele.valentini@mail.polimi.it |
Tool Description
Evolutionary Optimization Tool (Evoptool) is an open source optimization tool, distributed under GNU General Public License, implementing meta-heuristics based on the Evolutionary Algorithms paradigm, that aims to provide a common platform for the development and test of new algorithms, in order to facilitate the performance comparison activity. Evoptool offers a wide set of benchmark problems, from classical toy samples to more complex tasks, and a collection of algorithm implementations from Genetic Algorithms and Estimation of Distribution Algorithms paradigms. Evoptool is flexible, easy to extend, also with algorithms based on other approaches other from EAs.
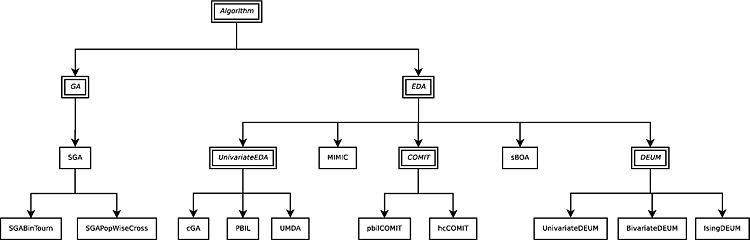
Algorithms
Evoptool offers a set of algorithm implementations coming from several paradigms within Evolutionary Computation, in the context of black-box optimization of boolean functions.
- Genetic Algorithms (GAs)
- Simple Genetic Algorithm (SGA). It is the original genetic algorithm with truncation selection, bitflip mutation, and single-point crossover.
- SGA with binary tournament selection. It is a variation of the SGA, and it differs for the selection operator that implements a binary tournament selection. Two individuals are picked up randomly from the population, and their fitness compared: the fittest individual is selected for the new population.
- SGA with uniform bitwise crossover. Another variation of SGA, it maintains the truncation selection and bitflip mutation, but implements a uniform bitwise crossover. This variation operator looks each bit of the two individuals string and swap them with a fixed crossover probability.
- Estimation of Distributions Algorithms (EDAs)
- Univariate EDAs:
- Population Based Incremental Learning (PBIL). It maintains a vector of marginal probabilities which is used to sample a new population at each generation. Then, after a selection of the population is performed, the marginal probabilities are computed again and the vector is updated according to a learning rule.
- Univariate Marginal Distribution Algorithm (UMDA). It differs from PBIL in having no learning rule. It simply generate new individuals using marginal probabilities computed as marginal frequencies of the previous population.
- Compact Genetic Algorithm (cGA). It is a space-efficient variation of PBIL that maintains a probability vector, but rather than generating a whole population, only two individuals are generated, from it and then are subjected to tournament. The probability vector is then updated according to a learning rule which considers both individuals.
- Univariate Distribution of Estimation Using Markov Random Field (Univariate DEUM). It uses an univariate Markov Random Field (MRF) to model the relationship between individuals and their fitness. MRF parameters are estimated through maximum likelihood, then the model is used to sample a new population by employing a Gibbs or Metropolis sampler.
- Bivariate EDAs:
- Mutual Information Maximizing Input Clustering (MIMIC). It adopts a directed conditional chain model for the distribution structure, learned by a greedy heuristic employing Shannon's Information Entropy, in order to minimize the Kullback-Leibler Divergence between the model and the true distribution.
- Combining Optimizers with Mutual Information Trees (COMIT). It is similar to MIMIC, but with a tree-structured Bayesian network rather than a single chain. A maximum weight spanning tree is used to learn the structure, guided by the entropy information principle. Once the structure is defined, it samples the individuals according to defined model, and then it exploits fast search algorithms to further improve the generated individuals. Inside Evoptool there are two implementation: one with an hill-climber fast search, and one with a PBIL based fast search.
- Bivariate Chain DEUM (Bivariate DEUM). It differs from the univariate version only for the structure of the underlying probabilistic model, that takes the form of a bivariate undirected chain.
- Ising Model DEUM (Ising DEUM). It is a DEUM like algorithm specifically designed to solve the Ising Spin Glass problem, by employing a bivariate lattice structure.
- Multivariate EDAs:
- Simple Bayesian Optimization Algorithm (sBOA). It uses a Bayesian network to represent the fitness structure, it employs the Bayesian-Dirichlet metric to measure the network quality, and a greedy algorithm to search the space of possible networks. At each generation, it computes and stores in the network the marginal and conditional probabilities of a selection of the original individuals, and than uses the Bayesian network to generate the new population.
- Univariate EDAs:
Benchmarks
Evoptool is designed to support the study of black-box optimization for boolean functions. The fitness function of each benchmark accepts as argument a string of boolean variables and returns a real value. Being performance comparison the main purpose of Evoptool, it already includes several optimization problems with different properties, varying from classical toy problems to more complex ones.
A first set of benchmarks is composed of simple toy problems without any variable dependency, therefore with an underlying univariate structure.
- One Max. A simple problem where the returned value is represented by the count of alleles of the individual taking value one. It has a unique optimal solution and every allele has the same relevance in the individual.
- One-Zero Max. It is a variation of One Max, where the fitness of an individual is represented by the maximum between the count of zeros and the count of ones. The relevant property of this problem is the existence of two optimal solutions: one with all alleles taking value one, and one with all zeros.
- Sum Value. In this toy problem the optimal solution is represented by a string of all ones. The fitness value is computed as the sum of the products between alleles and coefficients equal to the allele position inside the string. Also in this case there are no variable dependencies, but in contrast to the previous benchmarks the relevance of each allele is different and it grows linearly.
- Binary Value. Very similar to the Sum Value benchmark, it gives an increasing quadratic relevance at each alleles.
Another group of functions is constituted by deceptive problems, designed to mislead classical Genetic Algorithms. Their structure defines dependencies among variables with different orders (bivariate, trivariate, multivariate).
- Alternated Bits. Also called 1D Checkerboard, it introduces dependencies between couples of adjacent variables defining a chain structure. The problem takes into account the value of a variable relative to that of its immediate neighbours in the chain, in particular, higher fitness is achieved when neighbourring variables take opposite values.
- F3-Deceptive, Overlapping and Bipolar. This deceptive function is composed of separable building blocks of order three which has one global optimum for all ones and a deceptive local optimum to all zeros. The final fitness value is composed by the sum of the contributions of each deceptive pattern defined on three consecutive bits. The Bipolar variant is based on blocks of six bits, where there are two global optimum (all ones and all zeros) and a deceptive local optimum(three ones and three zeros). Overlapping version introduces overlaps among building blocks.
- Trap-5. This benchmark is similar to previous deceptive functions, and differs for the length of building blocks. As the previous functions, it has a global optimum for all alleles taking value one, and a deceptive local optimum for all zeros.
- Four Peaks and Six Peaks.
- Quadratic.
- Liepins Vose Fully Deceptive
Last but not least, there are implementation of real-like applications as:
- MAXSAT.
- Ising Spin Glass.
- Texture Restoration.
Statistics
Tool Usage
Evoptool is distributed under GNU General Public License and, so far, available for GNU/Linux platforms. Evoptool exploits several open source libraries and software, and it allows to wrap external algorithm implementations, as for Simple Bayesian Agorithm, reducing the efforts for the development. In this section, are described in the details the procedures to install the tool on your system.
Download
As metioned above, Evoptool implements several optimization algorithms, and it can also wrap external algorithm implementations through patches. This characteristic of the tool need to be correctly considered: if you plan to write an new algorithm then it is prefered to write it directly into Evoptool packages, if you just have the source code of an algorithm, and you want to save the time to implement it again, you can modify your code in order to work within Evoptool.
Description Version Date Link Evoptool with sBOA support. Evoptool without sBOA support. 1.0 24/02/2010 Evoptool1.0
External algorithm implementations cannot be directly distributed within Evoptool, but need to be downloaded from their original position, and, to be adjusted to work with Evoptool by applying a patch. Here you can find reference to wrapped algorithms and their patches to work within Evoptool.
Description External link Patch Simple Bayesian Algorithm sBOA
Requirements
In order to compile Evoptool, you need to install on your system some open source software and libraries, needed by the tool.
- Libraries:
- gtkmm-2.4
- glademm-2.4
- gthread-2.0
- sigc++-2.0
- cv
- highgui
- cvaux
- gsl
- gslcblas
- libxml++-2.6
- Software:
- gnuplot
Compilation
Evoptool source files and external algorithm implementations need to be compiled separately. In this section are described, step by step, all you need to compile the tool.
Evoptool package
If you want to compile the entire package, including the whole set of Evoptool libraries, you need to:
- In the main folder ../evoptool/trunk/ give a make cleanall command in order to remove all the binary files and library dependencies, and be sure to compile the entire project in the right way.
- In the main folder ../evoptool/trunk/ give a make all command in order to compile all the libraries and build the executable file.
If you need only to compile a single library, you need to:
- In the folder ../evoptool/trunk/mylib/src/ give a make lib command in order to compile and build library associated with that module. Repeat this operation for each library you want to compile.
- In the folder ../evoptool/trunk/gui/src give a make exe command in order to build the executable file.